Abstract
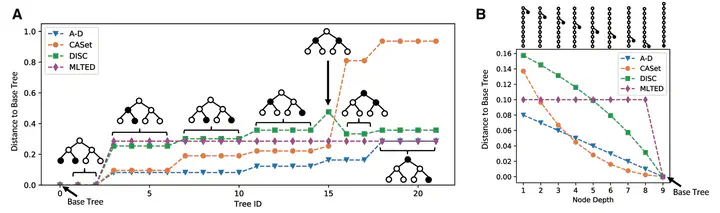
There has been recent increased interest in using algorithmic methods to infer the evolutionary tree underlying the developmental history of a tumor. Quantitative measures that compare such trees are vital to a num- ber of different applications including benchmarking tree inference methods and evaluating common inheritance patterns across patients. However, few appropriate distance measures exist, and those that do have low resolution for differentiating trees or do not fully account for the complex relationship between tree topology and the inherit- ance of the mutations labeling that topology. Here, we present two novel distance measures, Common Ancestor Set distance (CASet) and Distinctly Inherited Set Comparison distance (DISC), that are specifically designed to account for the subclonal mutation inheritance patterns characteristic of tumor evolutionary trees. We apply CASet and DISC to multiple simulated datasets and two breast cancer datasets and show that our distance measures allow for more nuanced and accurate delineation between tumor evolutionary trees than existing distance measures.