CS 321
Overlapping set of points
We consider two set of points: ![]() and
and ![]() that describes
two three-dimensional objects (e.g.,
that describes
two three-dimensional objects (e.g., ![]() of protein
chains). The (square of the) distance between the two coordinate sets is
defined by
of protein
chains). The (square of the) distance between the two coordinate sets is
defined by
![]() (1)
(1)
where ![]() are the
Cartesian position of one point (
are the
Cartesian position of one point (![]() coordinates).
coordinates).
We did not write ![]() but
but ![]() to indicate that
there is a complication. Each of the protein chains remains unchanged upon
rigid body translations and rotations. For example, if I will increase each of
the
to indicate that
there is a complication. Each of the protein chains remains unchanged upon
rigid body translations and rotations. For example, if I will increase each of
the ![]() by 5 the second
protein will still look exactly the same. More precisely, any transformation
that maintains the same distances between the points that make the object is a
rigid body transformation.
by 5 the second
protein will still look exactly the same. More precisely, any transformation
that maintains the same distances between the points that make the object is a
rigid body transformation.
- There is an exception to this case. What is it?
It is easy to verify that the displacement of all the ![]() by 5 did not
alter any of the distances. Consider the original
by 5 did not
alter any of the distances. Consider the original ![]() , it does not change by adding five to
, it does not change by adding five to ![]() and
and ![]() . That is,
. That is, ![]() . The conclusion is therefore that translations (three
translations along the x, y, and z axes) are legitimate rigid body
transformations. Similarly we can show that rotations do not change distances.
. The conclusion is therefore that translations (three
translations along the x, y, and z axes) are legitimate rigid body
transformations. Similarly we can show that rotations do not change distances.
Consider a position vector ![]() . A rotation is performed by a applying a matrix
. A rotation is performed by a applying a matrix ![]() on the vector
on the vector ![]() .
. ![]() satisfies
satisfies ![]() and
and ![]() . A distance between two points of the second object can be
written as:
. A distance between two points of the second object can be
written as:
![]()
If we apply the same rotation for every single vector representing point, the distance does not change:
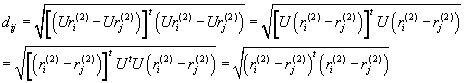
Translations and rotations do not affect the internal distances between internal points of a single object. However, if these operations apply to one object the distance between the two objects (equation (1)) depends on these operations. We need to make the definition of the distance unique. We therefore define the distance as the minimum value obtained by selecting optimal translations and the rotations.
For simplicity we consider separately the translations and the rotations. However, the separation does not affect the generality of our results.
Let us optimize the translation. We add to the points of the
first object a displacement vector ![]() . We have
. We have
![]()
We wish to determine ![]() so that
so that ![]() is a minimum.
Differentiating
is a minimum.
Differentiating ![]() with respect to
with respect to ![]() we obtain
we obtain
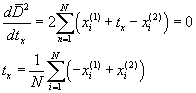
A similar argument follows for ![]() and
and ![]() .
.
Ok, that was easy. How about rotations? Rotations are a
little trickier; we will discuss them in steps. Let us first write the formula
for the distance in an adjusted form. We assume that the translations were
already taken care of (by fixing the geometric centers of the two proteins to
the (0,0,0)) coordinate) and apply the rotation for all points in the first
object. We also have to make sure that the matrix is unitary ![]() and for that
purpose we add Lagrange multipliers. We have (in an explicit form)
and for that
purpose we add Lagrange multipliers. We have (in an explicit form)
![]()
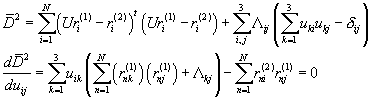
Consider the 3x3 matrices:
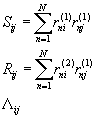
The minimum condition is written in a matrix notation as ![]() . Note that it is not so simple to solve the matrix e1quation
directly since we do not know
. Note that it is not so simple to solve the matrix e1quation
directly since we do not know ![]() and
and ![]() . In fact in many cases we can skip Alternatively multiplying the above equation by its
transpose, we have:
. In fact in many cases we can skip Alternatively multiplying the above equation by its
transpose, we have: ![]() . The matrix
. The matrix ![]() is symmetric but
is symmetric but
![]() is not.
is not.
Let ![]() be the
eigenvectors of
be the
eigenvectors of ![]() . They are also the right eigenvectors of
. They are also the right eigenvectors of ![]() . Let
. Let ![]() bet the left
eigenvector of
bet the left
eigenvector of ![]() ,
, ![]() is also the
product of applying
is also the
product of applying ![]() on
on ![]() (
(![]() ).
).
The eigenvalues of the symmetric matrix ![]() are
are ![]() .
.
Substituting in the expression for ![]() we have
we have
![]()
Clearly the first summation over the squares of the vectors
from the first and second objects is independent of the rotations. Note also
that in writing the expression in the second line we used the unitary property
of the rotation matrix (otherwise ![]() ). The rotation
matrix plays a role only in the last sum. Let
). The rotation
matrix plays a role only in the last sum. Let ![]() and
and ![]() be the left and
right unit vectors that form an orthonormal basis for the matrix
be the left and
right unit vectors that form an orthonormal basis for the matrix ![]() . That is
. That is ![]() or
or ![]() . We have now
for the last sum
. We have now
for the last sum
![]()
![]()
Suggest now an algorithm to compute the minimal distance.
Suggest a procedure to determine the rotation matrix.
Are we done, or are there more problems to fix?