CS 2110 Assignment 5
Due: Thu 12/3 11:59pm
This assignment builds on Assignment 4, and again you can work with a
partner.
Objectives
1.
We’ll write a recursive procedure that, given a
phylogenetic tree, outputs a file in the format used by the “Dendroscope”
program. This format is called “Extended
Newick tree format" and is a standard
that you can read about here. Please visit the Newick link and make sure
you understand the format. Name the file
“phylogeny.tre”.
2.
The particular tree we’ll create will be the one
generated by your solution to A4. Our
ultimate goal is to run Dendroscope to visualize the family tree of animals
A0-A39, using animal A38 as the root.
However you may want to work with a smaller set of animals, perhaps A0
to A3, until you get the hand of things.
3.
Using the GUI you developed for assignment 3, you’ll
also display the phylogeny relationships in a color-coded way.
4.
For those interested in competing in the CS 2110
competition, there will be an optional additional assignment involving the
automated discovery of the root node.
Required Portion
Building on Assignments 3
and 4
To tackle Assignment 5 you will need to start
with a working version of Assignment 4 and a working GUI from Assignment 3. We will not be providing a solution for
Assignment 4: use your own code.
Likewise, you can reuse the tests from previous
assignments. In fact, this is a good
strategy to catch regression bugs.
Regressions are bugs which break existing features as opposed to new
features. If you have good test
coverage, then you can run the tests frequently to protect against regressions
Dendroscope
The Dendroscope application is an existing tool
for displaying relationships between animals in various ways. You will need to download it from the web, at
this download link. You will need to request a key; follow the
instructions on the link page to do so.
Or if you just ignore it when it asks you for a key, that may also work.
Dendroscope generates a phylogeny tree by
reading from a file that ends in the extension .tre. It has a very
specific (and peculiar) format, the Newick file format. Here’s
an example :
([Fishus BallToothium]Toothy_Ballonfish:0.45, [Fishus
Paridisius]Paradise_Rockfish:0.717) [Herius Grenium]Green_Herring
As you can see, these trees are graphs where each node is labeled using
the Name and LatinName fields from the data file. This particular example creates a tree with
three nodes. The root node is the Green
Herring, and it has two children: the Toothy Balloonfish and the Paradise
Rockfish.
The general rule is that each node is
associated with an expression of the following form:
[LatinName]Name[:branch_length]
A node that has children is expressed this
way:
(child1, … childn)[LatinName]Name[:branch_length]
branch_length tells Dendroscope how long to make the edges in the graph – if you omit
it, it uses 1.0 as the default. We
omitted any edge-length for the Green Herring because it will be the root of
the tree: it has no parent, so there is no edge. The Toothy Balloonfish will be below the
Green Herring on an edge of length 0.45, and Paradise Rockfish will be placed
below the Green Herring on an edge of length 0.717.
When you run Dendroscope, you need to tell it to open the file you
created. It will then graph the tree
described by that file. If you wish,
you can then tell Dendroscope to “Load taxon images” under the “Options” menu. Select the file folder (directory) in which
the animal PNG or BMP files we gave you are stored. Dendroscope will match the animal names in
the tree with the animal pictures in the folder and should display pictures
next to the nodes in the tree.
Note: Dendroscope uses a rather complicated algorithm
to figure out which animal picture goes with which animal name. Even though our animals have identical names
to their picture files, it sometimes gets confused. So if you manage to produce a tree that has
all the nodes labeled correctly and yet the wrong pictures are displayed next
to those nodes, it isn’t your fault.
With the full 40-animal tree, Dendroscope seems to work correctly, at
least for us.
Here's an example of a Dendroscope tree (without taxon pictures) in various
display options it offers.
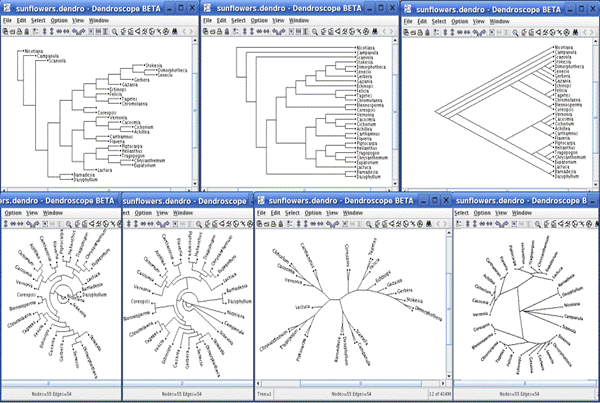
For our purposes in CS2110 we'll use the top left tree representation
but unlike the example shown, our trees will have animal names and images
associated with the "inner" nodes.
GUI
Use the GUI developed for assignment A3 to also
display data about the animals in a graphical, color-coded manner. If an animal is clicked, show that animal’s
descendents by highlighting them using background colors that indicate the
depth of each animal in the tree relative to the clicked animal.
For example, if you build a tree and then click
on the Spotted Salamander, only that animal and its descendants will be
highlighted. Immediate children will be
colored with one color, perhaps green, second level descendents will be colored
with a second color, perhaps red, etc.
Running the Program
Finally, to
run the program, provide a class which contains your main method. This class should not be the Animal class,
the PhylogenyTree class, one of the GUI classes, etc. It is just a separate class to start the
program. It will receive arguments
something like:
-cmd|-gui root *.dat out.tre
The first
argument is whether it should use the command line to create the Newick file or
the GUI to display the colored tree. Next comes the name of the root node, with no
extension (no “.dat”, for example “A18”).
The next arguments are the .dat
files to read in. If and only if you are
using -cmd option, the last argument is the name of the file to write the
Newick output to.
What to Submit
You will
submit this project exactly the same way you submitted it for A4. Thus, all of your classes (including the GUI
classes you write) should be in the phylogeny package or one of its
subpackages.
While any
package structure following this specification will be correct, you may lose
style points if you have an illogical package structure. For example, it would behoove you to put GUI
classes in a package called phylogeny.gui.
For
printing a tree in the Newick format, we’ll add a new method to the TreePrinter
class. Here is the signature you should
use:
/**
* Prints the phylogeny tree in the Newick format understood by Dendroscope.
* @param node - root node of tree
* @param stream - stream to print to
*/
public static void
printNewickFormat(PhylogenyTree node, PrintStream stream) {
// TODO
}
Also, you
can copy and paste this code into TreePrinterTests.java
/**
* Dummy test. Just prints the Newick format.
*/
@Test
public void testNewickDummy() throws
IOException {
ByteArrayOutputStream
out = new ByteArrayOutputStream();
TreePrinter.printNewickFormat(getPhylogenyTree(),
new PrintStream(out));
}
Note that the test
doesn’t actually “test” anything; you can only fail it by not conforming to the
specification or writing a printNewickFormat() functions which unexpectedly
throws an exception.
Note: You
shouldn’t submit any .jar files we provided for you (or their source
code). These would be the interfaces for
A1 as well as the GUI .jar in A3. You
can assume that we’ll have a copy of those and will automatically include them
in the classpath when compiling your code.
However, if you used an old version of the GUI library from A3, it
wouldn’t hurt to leave us a note in README.txt.
Helpful Note
Unless you
do the optional part, you shouldn’t ever have to touch the code for the Animal
or PhylogenyTree class. Now our design
decisions that we made in A4 should start to pay off!
You can now
modify the GUI as well as add a new function to print the tree in a Newick
format without modifying the animals or phylogeny tree itself. Separating the creation of data and the
display of data is a useful technique in software engineering. It’s very hard to cause regressions in the
animal or phylogeny tree if we don’t alter it!
Optional Portion: Awesome
Competition for Big Prizes!
Assignment 5 will also feature the annual competition to be the best
hacker in CS2110! We’ll have three prizes: Best solution, Best code quality and best quality assurance. (No team can win more than one prize). Winners will be announced at the final exam. The prizes will be gift certificates for $25
at Plum Tree (a Japanese restaurant in Collegetown).
To win the best solution prize, you
need to hit every button: good algorithms,
a clear writeup, excellent code, and thorough tests. The code quality prize and quality
assurance (testing) prize also require a good algorithm and a good
discussion, but our emphasis will be on other issues, such as the elegance of
your solution, the quality of comments, the quality of your JUnit testing, etc.
To put your program into this mode, specify “-chooseroot” instead of the root argument. For example, your program could be run with
arguments –gui –chooseroot A*.dat which
would run it on all files in the current directory that end in the .dat
extension, tell the application to use your automated root selection algorithm,
and run the GUI interface to visualize the result. The Newick file would not be created because
we are in GUI rather than CMD mode.
Here are some ideas for the competition, though you are certainly not
limited to these (if you really feel like writing a Fibonacci heap to build an
MST, we won’t stop you). Make sure you
do not alter the specification for a feature.
For example, if your program wants to take additional command line
arguments, it should still work as it usually does it you omit those extra
arguments.
Please submit a README.txt
telling us what you did and where we should look for the extra parts.
Finding the Root of the
Evolutionary Tree
Here’s what you need to do to compete.
Up to now, we’ve told you which animal should be at the root of the
evolutionary tree. ? Your challenge is to develop an algorithm for
discovering the best tree root in an automated
way that you can explain clearly. Hint: In thinking about your
algorithm, we recommend that you ask yourself what it takes to for an
evolutionary tree to “make sense”.
Another hint: you may also want to revisit the “hack” we used to push
animals away from their siblings. There
is something very clumsy about using the “shared gene” rule this way. Can you come up with a more elegant solution
in which we don’t have to modify the animal-to-animal distances computed back
in A3, where we didn’t use that shared-gene mechanism? We’re posing this as a genuine question: Ken,
at least, couldn’t come up with a better way to do it. But maybe you can do so, and if so, he’ll be
impressed! (Sushi, here we come....)
Once you have designed an algorithm, implement your solution and test it
on various data sets. It should look at
all the plausible roots for a given set of animals and select the one that
“wins” under the rules of your algorithm.
Obviously, it would surprise us if the right root for A0-A39 wasn’t
animal A38. But with some other set of
animals, the best choice of root might not be quite so easy to determine. We’ll test your solution on some unusual data
sets to see how you do… Good luck!
For uniformity, people who do the extra part of this assignment should
implement the following interface within their Phylogeny class:
Animal chooseRoot(Set<Animal> animals) {
...
}
Prim’s Algorithm
In A4, we
used a naïve algorithm to calculate the minimum spanning tree for our graph of
animal distances. Now we know that the
best algorithm to use is Prim’s. If you
decide to reimplement your solution, Java’s PriorityQueue class will be very
useful.
Graphviz
A very
useful program for displaying graphs can be downloaded for free. Graphviz reads in a file written with a special syntax
and creates a picture of the graph from it.
Using
Graphviz, create a visualization of the weighted, undirected graph of animal
distances (two siblings should have no edge instead of an infinite weight
edge). Furthermore, have this graph
display the minimum spanning tree.
Here is a
sample file used by Graphviz:
graph
G {
A[label="Turtle"];
B[label="Velociraptor"];
C[label="Penguin"];
A -- B [label="2"];
A -- C [label="1"];
B -- C [label="3",
style="dotted"];
}
Here is the graph produced by it.
Edges not in the MST are dotted. Feel free to play around with Graphviz to make
the graph look even better.
