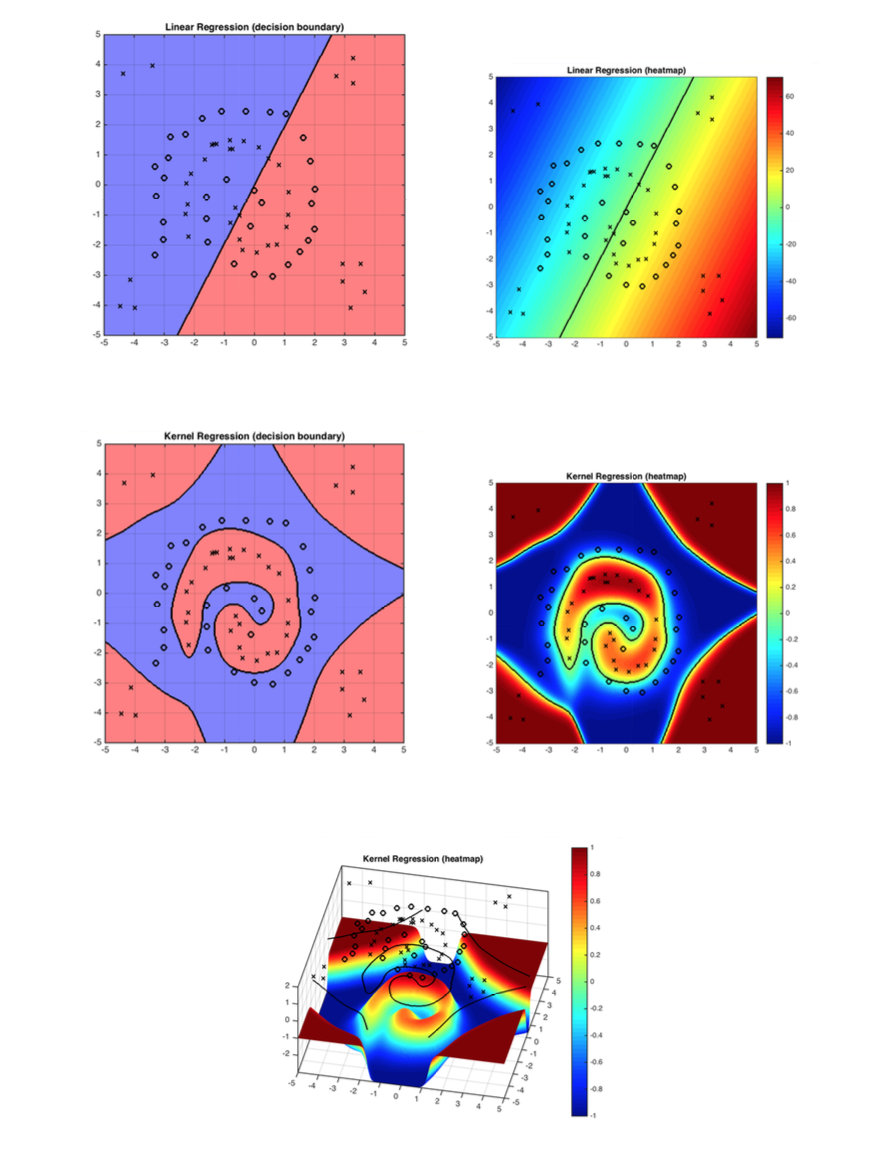
Figure 1: The demo shows how kernel function solves the problem linear classifiers can not solve. RBF works well with the decision boundary in this case.
Advantage: It is simple, and your problem stays convex and well behaved. (i.e. you can still use your original gradient descent code, just with the higher dimensional representation)
Disadvantage: \(\phi(\mathbf{x})\) might be very high dimensional.
Consider the following example: \(\mathbf{x}=\begin{pmatrix}x_1\\ x_2\\ \vdots \\ x_d \end{pmatrix}\), and define \(\phi(\mathbf{x})=\begin{pmatrix}1\\ x_1\\ \vdots \\x_d \\ x_1x_2 \\ \vdots \\ x_{d-1}x_d\\ \vdots \\x_1x_2\cdots x_d \end{pmatrix}\).
Quiz: What is the dimensionality of \(\phi(\mathbf{x})\)?
This new representation, \(\phi(\mathbf{x})\), is very expressive and allows for complicated non-linear decision boundaries - but the dimensionality is extremely high. This makes our algorithm unbearable (and quickly prohibitively) slow.
The kernel trick is a way to get around this dilemma by learning a function in the much higher dimensional space, without ever computing a single vector \(\phi(\mathbf{x})\) or ever computing the full vector \(\mathbf{w}\). It is a little magical.
It is based on the following observation: If we use gradient descent with any one of our standard loss functions, the gradient is a linear combination of the input samples. For example, let us take a look at the squared loss:
\begin{equation} \ell(\mathbf{w}) = \sum_{i=1}^n (\mathbf{w}^\top \mathbf{x}_i-y_i)^2\label{eq:c15:sql} \end{equation} The gradient descent rule, with step-size/learning-rate \(s>0\) (we denoted this as \(\alpha>0\) in our previous lectures), updates \(\mathbf{w}\) over time, \begin{equation} w_{t+1} \leftarrow w_t - s(\frac{\partial \ell}{\partial \mathbf{w}})\ \textrm{ where: } \frac{\partial \ell}{\partial \mathbf{w}}=\sum_{i=1}^n \underbrace{2(\mathbf{w}^\top \mathbf{x}_i-y_i)}_{\gamma_i\ :\ \textrm{function of \(\mathbf{x}_i, y_i\)}} \mathbf{x}_i = \sum_{i=1}^n\gamma_i \mathbf{x}_i \end{equation} We will now show that we can express \(\mathbf{w}\) as a linear combination of all input vectors, \begin{equation} \mathbf{w}=\sum_{i=1}^n \alpha_i {\mathbf{x}}_i.\label{eq:c15:alphas} \end{equation} Since the loss is convex, the final solution is independent of the initialization, and we can initialize \(\mathbf{w}^0\) to be whatever we want. For convenience, let us pick \(\mathbf{w}_0=\begin{pmatrix}0 \\ \vdots \\ 0\end{pmatrix}\). For this initial choice of \(\mathbf{w}_0\), the linear combination in \(\mathbf{w}=\sum_{i=1}^n \alpha_i {\mathbf{x}}_i\) is trivially \(\alpha_1=\dots=\alpha_n=0\). We now show that throughout the entire gradient descent optimization such coefficients \(\alpha_1,\dots,\alpha_n\) must always exist, as we can re-write the gradient updates entirely in terms of updating the \(\alpha_i\) coefficients:Formally, the argument is by induction. \(\mathbf{w}\) is trivially a linear combination of our training vectors for \(\mathbf{w}_0\) (base case). If we apply the inductive hypothesis for \(\mathbf{w}_t\) it follows for \(\mathbf{w}_{t+1}\).
The update-rule for \(\alpha_i^t\) is thus \begin{equation} \alpha_i^t=\alpha_i^{t-1}-s\gamma_i^{t-1}, \textrm{ and we have } \alpha_i^t=-s\sum_{r=0}^{t-1}\gamma_i^{r}. \end{equation} In other words, we can perform the entire gradient descent update rule without ever expressing \(\mathbf{w}\) explicitly. We just keep track of the \(n\) coefficients \(\alpha_1,\dots,\alpha_n\). Now that \(\mathbf{w}\) can be written as a linear combination of the training set, we can also express the inner-product of \(\mathbf{w}\) with any input \({\mathbf{x}}_i\) purely in terms of inner-products between training inputs: \begin{equation} \mathbf{w}^\top {\mathbf{x}}_j=\sum_{i=1}^n \alpha_i {\mathbf{x}}_i^\top{\mathbf{x}}_j.\nonumber \end{equation} Consequently, we can also re-write the squared-loss from \(\ell(\mathbf{w}) = \sum_{i=1}^n (\mathbf{w}^\top \mathbf{x}_i-y_i)^2\) entirely in terms of inner-product between training inputs: \begin{equation} \ell(\mathbf{\alpha}) = \sum_{i=1}^n \left(\sum_{j=1}^n\alpha_j\mathbf{x}_j^\top \mathbf{x}_i-y_i\right)^2\label{eq:c15:sql:ip} \end{equation} During test-time we also only need these coefficients to make a prediction on a test-input \(x_t\), and can write the entire classifier in terms of inner-products between the test point and training points: \begin{equation} h({\mathbf{x}}_t)=\mathbf{w}^\top {\mathbf{x}}_t=\sum_{j=1}^n\alpha_j{\mathbf{x}}_j^\top {\mathbf{x}}_t. \end{equation} Do you notice a theme? The only information we ever need in order to learn a hyper-plane classifier with the squared-loss is inner-products between all pairs of data vectors.Let's go back to the previous example, \(\phi(\mathbf{x})=\begin{pmatrix}1\\ x_1\\ \vdots \\x_d \\ x_1x_2 \\ \vdots \\ x_{d-1}x_d\\ \vdots \\x_1x_2\cdots x_d \end{pmatrix}\).
The inner product \(\phi(\mathbf{x})^\top \phi(\mathbf{z})\) can be formulated as: \begin{equation} \phi(\mathbf{x})^\top \phi(\mathbf{z})=1\cdot 1+x_1z_1+x_2z_2+\cdots +x_1x_2z_1z_2+ \cdots +x_1\cdots x_dz_1\cdots z_d=\prod_{k=1}^d(1+x_kz_k)\text{.}\label{eq:c15:poly} \end{equation} The sum of \(2^d\) terms becomes the product of \(d\) terms. We can compute the inner-product from the above formula in time \(O(d)\) instead of \(O(2^d)\)! We define the function \begin{equation} \underbrace{\mathsf{k}(\mathbf{x}_i,\mathbf{x}_j)}_{\text{this is called the} \textbf{ kernel function}}=\phi(\mathbf{x}_i)^\top \phi(\mathbf{x}_j). \end{equation} With a finite training set of \(n\) samples, inner products are often pre-computed and stored in a Kernel Matrix: \begin{equation} \mathsf{K}_{ij}=\phi(\mathbf{x}_i)^\top \phi(\mathbf{x}_j). \end{equation} If we store the matrix \(\mathsf{K}\), we only need to do simple inner-product look-ups and low-dimensional computations throughout the gradient descent algorithm. The final classifier becomes: \begin{equation} h(\mathbf{x}_t)=\sum_{j=1}^n\alpha_j\mathsf{k}(\mathbf{x}_j,\mathbf{x}_t). \end{equation}
During training in the new high dimensional space of \(\phi(\mathbf{x})\) we want to compute \(\gamma_i\) through kernels, without ever computing any \(\phi(\mathbf{x}_i)\) or even \(\mathbf{w}\). We previously established that \(\mathbf{w}=\sum_{j=1}^n\alpha_j \phi(\mathbf{x}_j)\), and \(\gamma_i=2(\mathbf{w}^\top \phi(\mathbf{x}_i)-y_i)\). It follows that \(\gamma_i=2(\sum_{j=1}^n \alpha_jK_{ij})-y_i)\). The gradient update in iteration \(t+1\) becomes $$\alpha_i^{t+1}\leftarrow \alpha_i^t-2s(\sum_{j=1}^n \alpha_j^tK_{ij})-y_i).$$ As we have \(n\) such updates to do, the amount of work per gradient update in the transformed space is \(O(n^2)\) --- far better than \(O(2^d)\).
Linear: \(\mathsf{K}(\mathbf{x},\mathbf{z})=\mathbf{x}^\top \mathbf{z}\).
(The linear kernel is equivalent to just using a good old linear classifier - but it can be faster to use a kernel matrix if the dimensionality \(d\) of the data is high.)
Polynomial: \(\mathsf{K}(\mathbf{x},\mathbf{z})=(1+\mathbf{x}^\top \mathbf{z})^d\).
Radial Basis Function (RBF) (aka Gaussian Kernel): \(\mathsf{K}(\mathbf{x},\mathbf{z})= e^\frac{-\|\mathbf{x}-\mathbf{z}\|^2}{\sigma^2}\).
The RBF kernel is the most popular Kernel! It is a Universal approximator!! Its corresponding feature vector is infinite dimensional and cannot be computed. However, very effective low dimensional approximations exist (see this paper).
Exponential Kernel: \(\mathsf{K}(\mathbf{x},\mathbf{z})= e^\frac{-\| \mathbf{x}-\mathbf{z}\|}{2\sigma^2}\)
Laplacian Kernel: \(\mathsf{K}(\mathbf{x},\mathbf{z})= e^\frac{-| \mathbf{x}-\mathbf{z}|}{\sigma}\)
Sigmoid Kernel: \(\mathsf{K}(\mathbf{x},\mathbf{z})=\tanh(\mathbf{a}\mathbf{x}^\top + c)\)
Can any function \(\mathsf{K}(\cdot,\cdot)\rightarrow{\mathcal{R}}\) be used as a kernel?
No, the matrix \(\mathsf{K}(\mathbf{x}_i,\mathbf{x}_j)\) has to correspond to real inner-products after some transformation \({\mathbf{x}}\rightarrow \phi({\mathbf{x}})\). This is the case if and only if \(\mathsf{K}\) is positive semi-definite.Remember \(\mathsf{K}_{ij}=\phi(\mathbf{x}_i)^\top \phi(\mathbf{x}_j)\). So \(\mathsf{K}=\Phi^\top\Phi\), where \(\Phi=[\phi(\mathbf{x}_1),\dots,\phi(\mathbf{x}_n)]\). It follows that \(\mathsf{K}\) is p.s.d., because \(\mathbf{q}^\top\mathsf{K}\mathbf{q}=(\Phi^\top \mathbf{q})^2\geq 0\). Inversely, if any matrix \(\mathbf{A}\) is p.s.d., it can be decomposed as \(A=\Phi^\top\Phi\) for some realization of \(\Phi\).
You can even define kernels over sets, strings, graphs and molecules.

Figure 1: The demo shows how kernel function solves the problem linear classifiers can not solve. RBF works well with the decision boundary in this case.