CS 2110 Homework Assignment 1. Due: Thu 9/9 11:59pm
This assignment is the first in a series that will span the semester. If you haven’t already, read about the complete project . The theme is computational biology: specifically, analyzing and comparing the genomes of a diverse set of species. We have provided the biological data in the SpeciesData.zip. The first step of the project is to read that data into our Java program.
But first, a word on style: Before you begin coding, you should familiarize yourself with the CS 2110 code style guidelines.
Download and install the JDK and Eclipse, if you don’t have them installed already. We recommend the latest stable release of each. Create a new Eclipse project using the Java Project wizard. Download and unpack the SpeciesData.zip archive, and move the SpeciesData folder into the top level of your Eclipse project. You can do this by dragging the folder’s icon onto the top-level icon in Eclipse’s “Package Explorer” pane. The data files have names like A12.dat and the images have names like Fuzzy_Tribble.png.
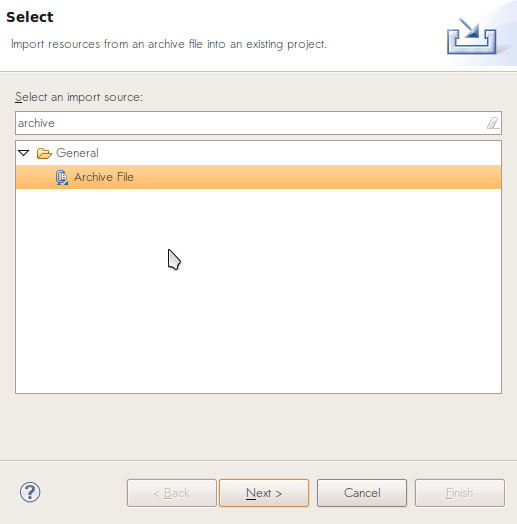
We’ve provided a source jar, a1.jar, to get you started. Source jars contain both compiled Java classes and source code. After downloading a1.jar, import its source code by selecting File -> Import and choosing “Archive File”.

Now you should now have four files in a new cs2110.assignment1 package:
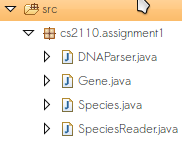
Do not modify these files.
Now, create your own package. Name it cs2110.netid.assignment1, where netid is your NetID. This is where your original source files will reside. We will follow this naming convention for later assignments.
The goals of this assignment are to read data from the species files and to parse their genomes. For each class in cs2110.assignment1, create a corresponding class in your package: MyDNAParser, MyGene, MySpecies, and MySpeciesReader. Each class should extend its namesake (or implement, in the case of DNAParser). Gene, Species, and SpeciesReader are all abstract classes, meaning that you will need to implement certain methods before a subclass can be instantiated. Hint: Let Eclipse fill in boilerplate code for superclass and interface methods that must be implemented.
Refer to the cs2110.assignment1 JavaDocs to learn more about these classes and the methods you will have to implement.
|
Name=”Biscuit” LatinName=”Retrieverus Aurum” ImageFilename=”Biscuit.png” DNA=”ITAYATYITITIAAYI” |
|
Your MySpeciesReader class is responsible for reading these files. The .dat files are text files, with one Attribute=”Value” pair on each line. You will parse these lines and set the corresponding variables in a MySpecies object.
Be sure to close the file when the EOF is reached.
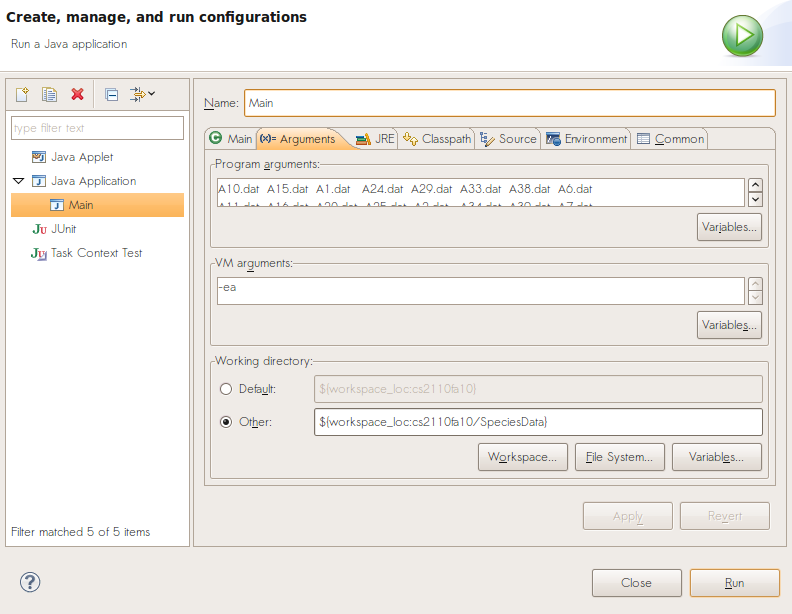
Also, do keep in mind that for an unqualified file pathname such as we see here, your program will look for the file in the place where it gets executed. From Eclipse, you control that choice of running directory from a form called the "Run Configurations..." on the "Run" pull-down tab. You can also specify the arguments to the Main method of your program there. Do not assume that data files are in any particular directory - it’s very bad form to hard-code a directory or file path.
DNA is a string of the characters {I,T,Y,A}. Genes are substrings of DNA, starting the special sequence IAY and terminated by TYI. Regions of DNA that are not in a gene are called non-coding, and can be discarded. Note that IAY can appear inside a gene, and TYI can appear in the junk DNA region. The IAY and TYI are not part of the gene.
See the DNAParser documentation for more information.
The Species class has a method getGenome with return type Collection<Gene>.
Important Note: The DNAParser might return duplicate genes, but getGenome() should return a collection that does not contain duplicates. A Set (such as HashSet) would be ideal for this application.
These four classes you’re implementing don’t do anything. To make things more interesting, we have suppied a main class that you can use to exercise your code: Main.java. Use of this class for testing is optional (and will not be graded), but highly recommended. Note that this program does not test the correctness of your output.
Copy and paste Main.java into your package and edit it to reflect your package and class names. Create a Run Configuration and list one or more .dat filenames as the program arguments. We recommend using SpeciesData as your working directory, so that you can refer to species data files without prefacing them with a relative path.
Eclipse is an enormously powerful tool that can help you complete this assignment in much less time. Some people are afraid of the initial investment it takes to learn, but after you do it's hard to believe you ever went without it. A few useful operations:
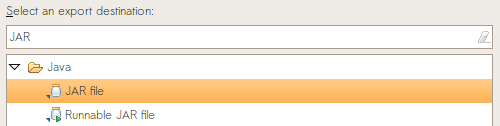
You will submit two files to CMS. The first is a source jar of your entire package to CMS containing your source code and compiled classes. To build a source jar, choose File -> Export, with “JAR file” as the export destination.
In the dialog that follows, it is very important that the box “Export Java source files and resources” is checked - otherwise, we won’t have any source code to grade.
After you have confirmed that all of your cs2110.netid.assignment1 package’s contents are checked, click “Finish” to finish exporting the jar. It’s a good idea to open your jar using your favorite ZIP file browser, just to double-check that your source code is included.
The second file is a write-up named README, in either text or PDF format. Briefly describe your experience working on the assignment: Any problems you faced and how you overcame them, what you learned, etc.
For a few extra credit points,
Note: We track extra credit as a separate column in CMS, so extra credit points will not be added directly to your assignment score. At the end of the semester, a stellar cumulative extra credit score could nudge a grade up by half a letter, e.g., from B+ to A-.